If file exists in folder read it else skip the processing part
Categories:
Conditional File Processing in R: Read if Exists, Skip Otherwise
Learn how to robustly check for file existence in R and conditionally read data, preventing errors and streamlining your data processing workflows.
In data analysis and scripting, it's common to encounter scenarios where you need to process a file only if it exists. Attempting to read a non-existent file in R will typically result in an error, halting your script. This article demonstrates how to implement a robust check for file existence before proceeding with file operations like read.table, ensuring your scripts run smoothly and handle missing files gracefully.
The Importance of File Existence Checks
Before performing any operation on a file, especially reading its contents, it's crucial to verify that the file actually exists at the specified path. Without this check, your R script might terminate unexpectedly with an error message such as cannot open file 'non_existent_file.txt': No such file or directory. Implementing a conditional check allows your script to either proceed with processing or skip it, log a message, or take an alternative action, making your code more resilient and user-friendly.
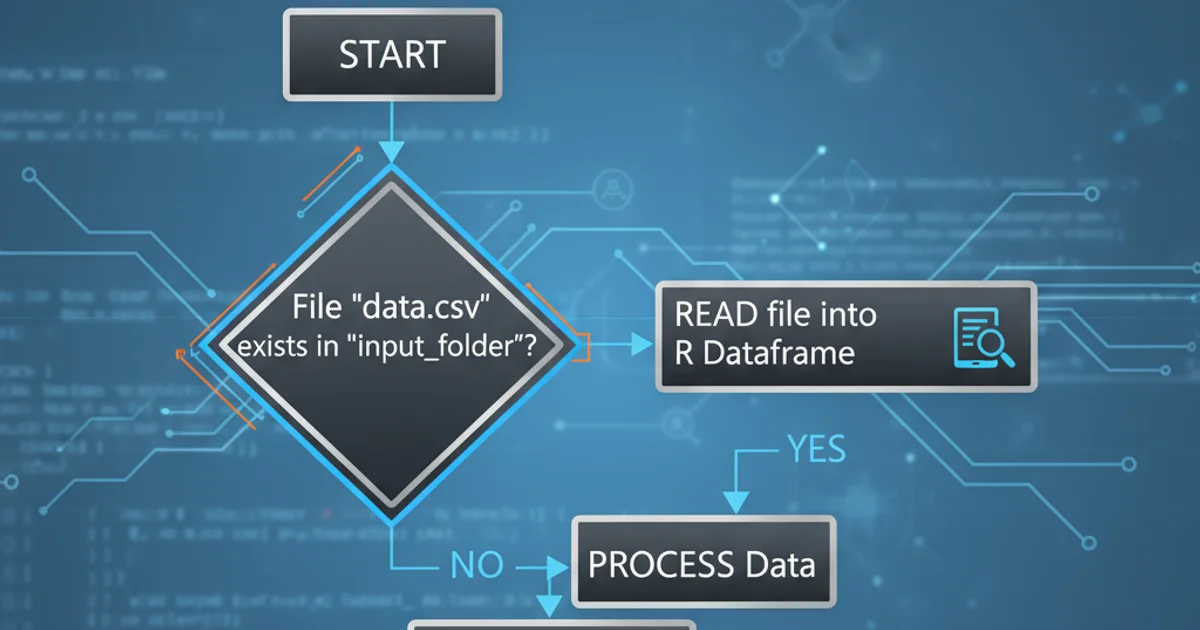
flowchart TD
A[Start Script] --> B{Does 'data.csv' exist?}
B -->|Yes| C[Read 'data.csv' into DataFrame]
C --> D[Process DataFrame]
B -->|No| E[Skip Processing]
E --> F[Log 'File not found' message]
D --> G[End Script]
F --> GWorkflow for conditionally reading a file based on its existence.
Implementing File Existence Checks in R
R provides the file.exists() function, which is specifically designed for this purpose. It takes a file path as an argument and returns TRUE if the file exists and FALSE otherwise. This function can be seamlessly integrated into an if statement to control the flow of your script.
# Define the file path
file_path <- "my_data.csv"
# Check if the file exists
if (file.exists(file_path)) {
# If the file exists, read it
my_data <- read.csv(file_path)
message(paste("File '", file_path, "' found and loaded.", sep=""))
# Perform further processing on my_data
print(head(my_data))
} else {
# If the file does not exist, skip processing and print a message
message(paste("File '", file_path, "' not found. Skipping processing.", sep=""))
}
Basic R script to check for file existence and conditionally read a CSV file.
read.csv() for comma-separated values and read.table() for space or tab-separated files. Adjust the sep argument in read.table() if your delimiter is different.Handling Multiple Files and Advanced Scenarios
For scenarios involving multiple files, you can iterate through a list of file paths and apply the file.exists() check within a loop. This approach is particularly useful when dealing with batches of data files that might not all be present. You can also combine file.exists() with error handling mechanisms like tryCatch for more sophisticated error management, though a simple if statement is often sufficient for basic existence checks.
# List of files to process
file_list <- c("data_day1.txt", "data_day2.txt", "non_existent_data.txt", "data_day3.txt")
# Loop through each file
for (file_name in file_list) {
if (file.exists(file_name)) {
message(paste("Processing file: ", file_name, sep=""))
# Example: Read and print first few lines
df <- read.table(file_name, header = TRUE)
print(head(df))
} else {
message(paste("Skipping file: ", file_name, " (not found)", sep=""))
}
}
Iterating through a list of files and conditionally processing each one.
normalizePath() or here::here() can help manage paths robustly.